Investigating the roles of Pol Epsilon in genome and epigenome stability
Primary supervisor: Roberto Bellelli, Queen Mary University of London
Secondary supervisor: David Michod, UCL
Project
Genomic instability is a major hallmark of cancer and understanding its nature has provided avenues for patient-tailored therapies. Recent studies uncovered DNA Polymerase Epsilon (Pol Epsilon), as a novel therapeutic target in cancer (1). Furthermore, genome wide screenings have discovered that deletion of the POLE3-POLE4 subunits of Pol Epsilon leads to increased sensitivity to ATR and PARP inhibitors (2, 3). The aim of this project is to unveil the mechanisms behind these observations to exploit Pol Epsilon, and POLE3-POLE4, as markers of sensitivity and novel therapeutic targets. The project will combine approaches of biochemistry and structural biology, cell biology and bioinformatics, available within the CRUK City of London network, to accomplish three aims:
Aim 1. Reconstitution of DNA replication and histone binding by Pol Epsilon.
We previously discovered that POLE3-POLE4 bind double stranded DNA (4); This activity might promote Pol Epsilon processivity and/or fidelity, thus preventing gap accumulation and/or nucleotide misincorporation at the replication fork. Loss of this function might then result in synthetic lethality with inhibition of PARP/ATR and the selective vulnerability of specific cancers (1). To address this hypothesis, we will investigate in vitro Pol Epsilon-dependent DNA synthesis in the presence/absence of POLE3-POLE4 (Figure 1). We also previously found that POLE3-POLE4 bind to histone H3-H4. To determine the functional relevance of this activity, we will investigate this interaction within full Pol Epsilon and identify the specific domains involved by a combination of structural biology approaches (including CRYO-EM in collaboration with Dr Cherepanov). This will allow the generation of separation of function mutants to investigate the role of Pol Epsilon histone binding in genome stability and sensitization to ATR/PARP inhibitors.
Aim 2. Dissection of POLE3-POLE4 functions and synthetic lethality with ATR/PARP inhibitors
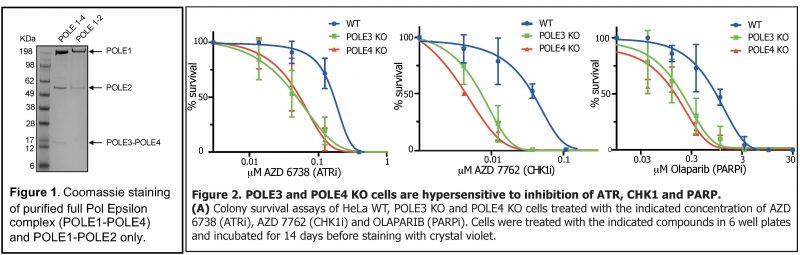
We recently showed an increased sensitivity of POLE3-POLE4 KO cells to ATR and PARP inhibitors (Figure 2). To understand the mechanism behind this observation, we will analyse the dynamics of DNA replication in POLE3/POLE4 KO cells in the presence of ATR and PARP inhibitors by DNA fiber assay (5). We will also analyse markers of replication stress and DNA damage (e.g. RPA, gH2AX and 53BP1) by confocal microscopy. Finally, we will employ CUT&TAG coupled to BrdU-IP, a technique we recently established with Dr Michod, to study the role played by the histone chaperone activity of Pol Epsilon in this phenomenon.
Aim 3. Analysis of Pol Epsilon expression in cancer.
In collaboration with bioinformatics at UCL, we will interrogate cancer databases for mRNA and protein expression levels of Pol Epsilon subunits in cancer and correlate it with survival, tumour features and mutational signatures. In addition, we will assess the expression levels of Pol Epsilon subunits in cancer cell lines presenting diverse sensitivity to PARP and ATR inhibitors. Importantly, we have already generated all the tools (e.g. specific antibodies) required to test the expression of these proteins and can access a major biobanks of cancer tissues at Barts Cancer Institute/ Queen Mary University of London.
Candidate background
This project would suit candidates with a background in biochemistry and cell biology and an interest in genome stability.
Potential Research Placements
- David Michod, Great Ormond Institute of Child Health/ UCL
- Peter Cherepanov, Francis Crick Institute
- Nnenna Kanu, UCL Cancer Institute
References
- Sviderskiy VO, Blumenberg L, Gorodetsky E, Karakousi TR, Hirsh N, Alvarez SW, Terzi EM, Kaparos E, Whiten GC, Ssebyala S, Tonzi P, Mir H, Neel BG, Huang TT, Adams S, Ruggles KV, Possemato R. Hyperactive CDK2 Activity in Basal-like Breast Cancer Imposes a Genome Integrity Liability that Can Be Exploited by Targeting DNA Polymerase ε. Mol Cell 80, 682-698.
- Hustedt N, Álvarez-Quilón A, McEwan A, Yuan JY, Cho T, Koob L, Hart T, Durocher D. A consensus set of genetic vulnerabilities to ATR inhibition. Open Biol. 9, 190156.
- Su D, Feng X, Colic M, Wang Y, Zhang C, Wang C, Tang M, Hart T, Chen J. CRISPR/CAS9-based DNA damage response screens reveal gene-drug interactions. DNA Repair (Amst). 87, 102803
- Bellelli R, Belan O, Pye VE, Clement C, Maslen SL, Skehel JM, Cherepanov P, Almouzni G, Boulton SJ. POLE3-POLE4 Is a Histone H3-H4 Chaperone that Maintains Chromatin Integrity during DNA Replication. Mol. Cell 72,112-126.
- Bellelli R*, Borel V*, Logan C, Svendsen J, Cox DE, Nye E, Metcalfe K, O’Connell SM, Stamp G, Flynn HR, Snijders AP, Lassailly F, Jackson A, Boulton SJ. Polε Instability Drives Replication Stress, Abnormal Development, and Tumorigenesis. Mol. Cell 70, 707-721. *equal contribution
