Mechanochemical control of nuclear architecture and drug resistance in breast cancer
Primary supervisor: Maddy Parsons, King’s College London
Secondary supervisor: Louise Jones, Queen Mary University of London
Project
Breast cancer is the leading cause of cancer mortality in women globally, with >650,000 deaths per year. Drug-resistant breast cancer remains a significant challenge, often occurring without clear genetic drivers and leading to disease progression and poor outcomes. Dynamic control of the actin cytoskeleton is essential for cancer cell division, pro-survival signalling and invasion. Cytoplasmic actin has been widely studied in cancer cells; however, functions of actin are not limited to the cytoplasm. Transient nuclear F-actin assembles post-mitosis in healthy cells and is proposed to provide a scaffolding function to organise chromatin, with potential roles in DNA repair. Roles for nuclear F-actin in cancer progression remain largely unexplored.
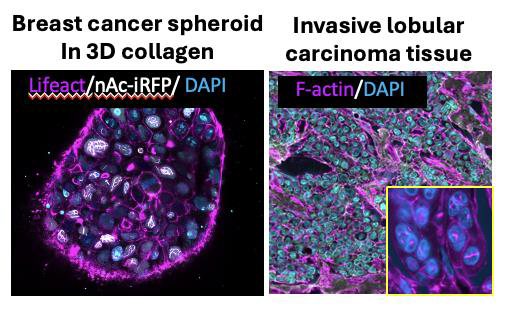
Our recent work used high content screening to reveal that nuclear F-actin scaffolds to histoneH3 to organise chromatin and control DNA repair in cancer cells [1]. We have also shown that mechanical forces and fibrotic extracellular matrix leads to pro-invasive signalling and reorganisation of cytoplasmic F-actin in 3D models of breast cancer [2,3]. Our unpublished data has further revealed that nuclear F-actin is upregulated in cancer spheroids and organoids in stiffer matrix (Figure, left panel), resulting in stiffer nuclei and protection from chemotherapy. Moreover, nuclear F-actin is absent from epithelial cells in normal human breast tissue but upregulated in invasive lobular breast carcinoma tissue (Figure, right panel). Recent evidence from our collaborators also shows that drug-resistant breast tumours stiffen one week after initiation of chemotherapy [4] and this persists during the ensuing ~6-month treatment period, indicating that rapid adaptation of tumour biomechanics is an inherent feature of non-responder patients. This combined evidence points towards a role for nuclear F-actin in controlling nuclear architecture and drug resistance in breast cancer.
The goal of this project is to define contributions from the evolving tumour microenvironment to nuclear F-actin, chromatin architecture and transcriptional response to chemotherapy. Advanced imaging and spatial omics analysis of human breast cancer samples coupled with 3D models of human breast cancer will enable identification of key features in human disease that contribute to nuclear F-actin assembly, structure, persistence and drug resistance. Building from our existing strong foundations and advanced computational pipelines, the project will drive a holistic understanding of the pathological features of aggressive breast cancer tissues that correlate with nuclear F-actin assembly, considering spatial extracellular matrix type, quantity, organisation; tumour-immune interactions; spatial single cell transcriptional signatures and tumour/nuclear architecture. Advanced breast cancer spheroid and organoid models in 3D matrices of differing stiffness, biophysical analysis and imaging evolution of tumour and stromal compartments will provide a foundation for perturbation of key identified pathways from human tissue to evaluate functional response. The project will also explore concepts of mechanical ‘memory’ in cancer progression and the contributions of nuclear F-actin to this process. The student will develop cross-disciplinary skills in cancer cell biology, pathology, imaging, bioinformatics and omics interpretation and computational integration. The outcomes of the project will inform on novel molecular mechanisms of drug resistance in breast cancer, identify potential biomarkers and features of tissues that predict resistance and potential therapeutic targets for intervention in future.
Candidate background
This project would be ideal for students with a degree/masters in life sciences, with a strong grounding in cancer cell biology, and interests in imaging-based spatial biology approaches, biophysics and computational analysis.
Potential Research Placements
- Anita Grigoriadis, PharosAI
- Robert Prevedel, EMBL, Heidelberg (D)
- Louise Jones, Barts Cancer Institute, Queen Mary University of London
References
- Lawson C, Peel S, Jayo A, Corrigan A, Iyer P, Baxter Dalrymple M, Marsh RJ, Cox S, van Audenhove I, Gettemans J, Parsons M. Nuclear fascin regulates cancer cell survival. 2022. eLife. doi: 10.7554/eLife.79283
- Hayward MK, Allen MD, Gomm JJ, Goulding I, Thompson CL, Knight MM, Marshall JF, Jones JL. Npj Breast Cancer. Doi: 10.1038/s41523-022-00464-4
- Jahin I, Phillips TA, Marcotti S, Gorey MA, Cox S, Parsons M. Extracellular matrix stiffness activates mechanosensitive signals but limits breast cancer cell spheroid proliferation and invasion. 2023. Front. Cell Dev Biol. doi: 10.3389/fcell.2023.1292775
- Sinha A et al, Tumor Biomechanics Quantified Using MR Elastography to Predict Response to Neoadjuvant Chemotherapy in Individuals with Breast Cancer. Radiology. doi: 10.1148/rycan.240138
