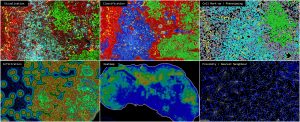
Tissue Cyclic Immunofluorescence and Analysis Core
Mapping the spatial organisation of tissues has proven to be an essential vantage point from which to investigate the cellular and signalling complexities that underly disease progression and therapeutic response within the tumour microenvironment. Through the integration of high dimensionality images, spatially resolved multi-omics has allowed for the simultaneous assessment of cellular function, local communication networks and whole tissue signature / state.
Tissue-cyclic fluorescence (T-CyCIF) describes a public domain, bench-based method for the acquisition of these highly multiplexed (high dimensionality) images. Through re-iterative staining cycles based upon conventional immunofluorescence reagents and microscopes, this approach is easily integrated into existing histopathological workflows for “limitless” 60 + marker interrogation.
T-CyCIF has witnessed a growing ecosystem of specialised platforms and end-to-end methods in recent years, each conveying relative advantages and disadvantages based on their application, practical demands and resource investment. Principally, the core service functions to support and develop T-CyCIF projects based upon (but not exclusive to) the Cell DIVE imager and PhenoCycler (CODEX®) platforms.


Contact
Academic Lead
Technologies
- Semi-Automated Immunofluorescence Imaging Platform.
- High throughput – 5 channels / 20x / 900mm2 / 165mins.
- Cyclic step staining (4 markers per round). 60+ targets.
- Disruptive photooxidative signal removal. Round Optimisation
- Open reagent system.
- Automated post processing (stitching + BG subtraction)
- High practical demands.
- Automated DNA Barcoding platform.
- High throughput – 4 channels / 40x / 225mm2 / 45mins
- Single step staining (master mix). 100+ targets.
- Non-destructive (de)hybridisation led signal removal.
- Expensive propriety oligonucleotide conjugations and reagents. Off-the-shelf Panels
- Automated post processing (stitching + BG subtraction)
- Moderate but specialised practical demands.
Services
- Optimised 1° Ab-fluorophore Information Sheet
- Panel Design and 1° Ab Optimisation Strategy Support
- T-CyCIF Protocol Demonstrations and Training
- Project Consultancy and Support
Contact: j.hartlebury@qmul.ac.uk
BCI HALO AI Facility
HALO AI is a comprehensive analysis software package with an intuitive user interface, applying AI driven deep learning networks for segmentation, classification, and phenotyping towards quantitative and spatial analyses.
- Non-proprietary (JPG, TIF)
- 3D Histech (MRXS)
- Akoya Biosciences (QPTIFF, component TIFF)
- Aperio (SVS, AFI)
- Leica (SCN, LIF)
- DICOM (DCM)
- Open Microscopy (OME.TIFF)
- Nikon (ND2)
- Olympus/Evident (VSI)
- Hamamatsu (NDPI,NDPIS)
- Zeiss (CZI)
- Vectana (BIF)
- KFBio (KFB)
- Philips (ISYNTAX, I2SYNTAX)
- Highplex FL – Incorporate “limitless” FL channels / markers into tissue overlay for interrogation of high-plex data.
- FISH / FISH-IF – Quantify FISH probe + IF detected marker expression on tissue.
- Multiplex IHC – Analyse brightfield images of up to 5-plex co-stains with deconvolution tool.
- ISH / ISH-IHC – Quantify ISH probe + chromogen detected marker expression.
- Spatial Analysis – Extension / plug-in module for all other modules, allows 1. Heatmaps 2. Nearest Neighbour 3. Proximity and 4. Infiltration analyses.
- TMA – Extension / plug-in module facilitating categorisation and separate analysis of TMA cores.
- Random Forest– Traditional Ensemble-method Machine Learning Tissue Classifier.
- Mininet– Deep Learning (AI) Tissue Mask Classifier for small sample training (Shallow).
- DenseNet V2 – Deep Learning (AI) Tissue Mask Classifier for large sample training (Robust).
- Nuclei Seg V2 (BF/FL 1.0.0)– Deep Learning (AI) Nuclei Classifier for Cell segmentation.
- Membrane Seg (BF/FL 1.0.0)– Deep Learning (AI) Membrane Classifier for Cell segmentation.
- Object Phenotyper– Deep Learning “train by example” tool to categorize objects based on morphology and/or biomarker expression.
- QC Slide – Pretrained DenseNet V2 Classifier chain consisting of a Tissue Detection Classifier and an Artifact Detection Classifier.
Services
- HALO AI Access
- General Enquiries
- Training
- Consultancy
Contact: j.hartlebury@qmul.ac.uk / s.wallis@qmul.ac.uk