Dissecting the roles of transposable elements in paediatric acute myeloid leukaemia
Primary supervisor: Özgen Deniz, Queen Mary University of London
Secondary supervisor: Richard Dillon, King’s College London
Project
Paediatric acute myeloid leukaemia (AML) is a highly heterogeneous disease that accounts for approximately 15- 20% of all childhood leukaemia cases. Despite the tremendous progress made over the past decades in diagnosis and treatment of paediatric AML, it remains a life-threatening cancer with an overall survival rate of ~65% (1). Recent genomic studies have shown remarkably few somatic mutations in paediatric AML compared to adult AML and other cancers (2), highlighting the need for alternative approaches to characterise potential targetable dependencies.
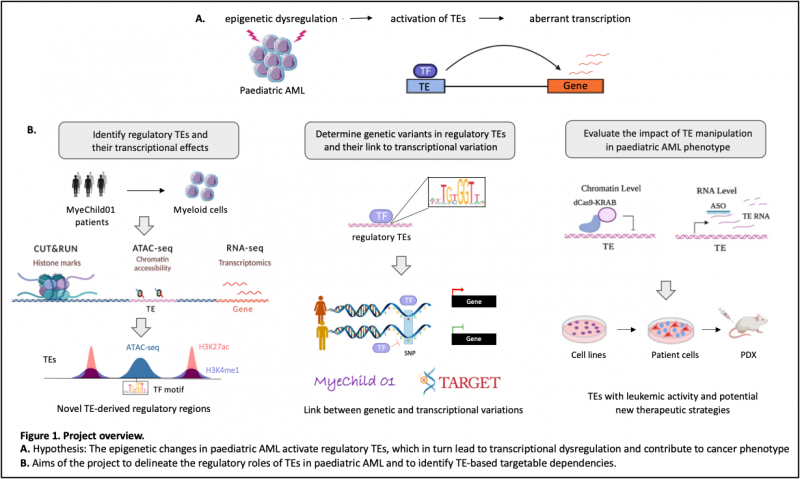
Initiation and progression of AML is often associated with dysregulation of the transcriptional machinery which is caused by alterations in the transcription factor (TF) activity and global epigenetic landscape. One less well characterised but very important source of transcriptional regulators are transposable elements (TEs). TEs are repetitive DNA sequences that compose half of our genome. Although most are usually repressed by epigenetic mechanisms, throughout evolution many TEs have been co-opted as gene regulatory regions and contribute up to 35% of TF binding sites (3). Their cryptic activation could therefore lead to dysregulation of regulatory and transcriptional networks in cancer. In fact, we previously demonstrated for the first time that TEs can be epigenetically activated and direct nearby gene expression in adult AML (4). However, these highly functional regions of the genome and their links to disease pathogenesis remain unexplored in paediatric AML.
The objective of this project is to delineate the molecular and cellular functions of TEs in paediatric AML and to identify TE-based targetable dependencies. To achieve this, we will combine epigenomic and transcriptomic assays with cutting-edge CRISPR-Cas9 genome editing and bioinformatics and implement innovative TE targeting strategies using antisense oligonucleotides. Importantly, this project will leverage the resources of clinical samples and data collected in the international MyeChild01 study, in which Dr Richard Dillon is one of the molecular leads. MyeChild01 completed recruitment of >700 children with AML and transcriptomic and genomic profiling have already been performed on diagnostic samples. The student will perform chromatin profiling of TEs in diagnostic samples using ATAC-seq and CUT&RUN methodologies and integrate these data with MyeChild01 transcriptomic data to identify regulatory TEs and their impact on transcriptional networks in paediatric AML. A potential link between inter-individual variations in regulatory TE sequences and gene expression among paediatric patients will be also tested using MyeChild01 as well as TARGET datasets (5). Finally, the student will assess whether targeting the identified regulatory TEs impacts cancer-specific gene expression patterns and reverses cancer cellular phenotypes in paediatric AML using in vitro, ex vivo and in vivo models.
This project will provide a comprehensive view on the roles of TEs as crucial transcriptional regulators in paediatric AML, which may lead to novel prognostic and therapeutic strategies to improve the survival of children with AML.
Candidate background
We seek highly motivated candidates with a background in cell and molecular biology and interest in cancer epigenetics. Experience in mammalian cell culture, CRISPR/Cas9 system and basic bioinformatic analyses is not required but would be advantageous.
Potential Research Placements
- Richard Dillon, King’s College London
- Juan Wang, Barts Cancer Institute/ Queen Mary University of London
- TBD: training in 3D culture models
References
- Pui, C.H. et al. (2011) Biology, risk stratification, and therapy of pediatric acute leukemias: An update. J Clin Oncol; 29:551-565.
- Radtke, I. et al. (2009) Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc Natl Acad Sci U S A; 106(31): 12944-12949.
- Chuong, E.B. et al. (2017) Regulatory activities of transposable elements: From conflicts to benefits. Nat Rev Genet; 18(2):71-86
- Deniz, O. et al. (2020) Endogenous retroviruses are a source of enhancers with oncogenic potential in acute myeloid leukaemia. Nat Commun.;11(1):1-14.
- Bolouri, H. et al. (2018) The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nature Medicine; 24, 103-112
