Testing the causal relationship between epigenetic drift, clonal haematopoiesis and malignant switch in human blood
Primary supervisor: Gabriella Ficz, Queen Mary University of London
Secondary supervisor: Dominique Bonnet, Francis Crick Institute
Project
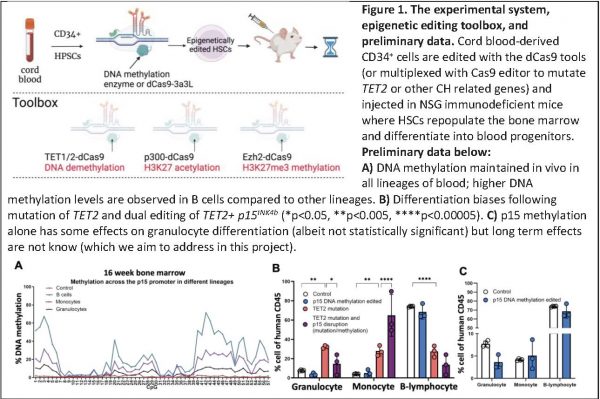
We hypothesised that DNA methylation hotspots observed in acute myeloid leukaemia (e.g. p15INK4b promoter hypermethylation) might happen early in the disease initiation process, and designed an experimental system to recapitulate such methylation events in normal haematopoietic/progenitor human stem cells (HSPCs). We have achieved, for the first time, robust epigenetic editing in HSPCs and can therefore measure deposition, maintenance and physiological impact of DNA methylation in vivo (unpublished work, see Figure 1). Epigenetic drift is apparent in functionally declining and ageing HSPCs, which also exhibit: a decreased self-renewal; decreased differentiation capacity; an increased myeloid bias with subsequent reduction in adaptive immune response; a reduced regenerative potential and clonal haematopoiesis (CH). CH is where distinct HSPCs, with or without a specific mutation, gain clonal advantage and expand to represent large fractions of the blood cell population, leading to disease incidence increase.
Notably, about a third of us will develop CH in old age. Two of the most common CH associated mutations (1) are those affecting DNMT3A and TET2, both enzymes involved in the control of DNA methylation in the genome. Strong associations between CH and accelerated epigenetic ageing have been recently identified (2), however causal relationship between DNA methylation CH and general physiological decline is unknown.
In this project we hypothesise that ageing associated epigenetic drift is causally linked to functional decline of human HSPCs leading to CH and malignancy development over time. Using CRISPR/dCas9-3a3L DNA methylation editing technology, in collaboration with the Bonnet lab, we find that ectopically edited HSPCs repopulate the bone marrow; maintain DNA methylation in both lymphoid and myeloid lineages of NSG mice for at least 16 weeks, and preliminary data indicate effects on differentiation, consistent with CH. However, the long-term effects of these changes are not known. We are also able to multiplex CRISPR/Cas9 editing to generate TET2 mutations and p15INK4b hypermethylation in the same HSPC. We are thus the first to demonstrate the feasibility of combining epigenetic and genetic editing in HSPCs and study the impact on HSPC physiology.
We are therefore in the unique position to combine state-of-the-art epigenetic editing with in vivo functional analysis of human HPSCs, and address the following fundamental questions in relation to the effects of epigenetic changes on DNA and histones, as seen in ageing and cancer:
- What are the long-term consequences of epigenetic changes on HSPC fate? What happens if regulatory sites, which control cellular identities, gain DNA and histone methylation stochastically as observed during ageing? What impact does inflammation have on survival, fitness and differentiation of epigenetically
edited cells? - What are the long-term consequences on clonal diversity in vivo? We will address this by combining epigenetic editing with Cas9 induced lineage tracing (3).
- Can we reverse the myeloid bias in HSPCs associated with DNA methylation (4)? We will aim to reverse these aberrations using targeted DNA demethylation catalyzed by TET dioxygenases (5).
These results will reveal the causal relationship between epigenetic drift and physiological HSPC decline, and will carve the technological development toward in vivo delivery of epigenetic editing tools to repair, inhibit and reverse ageing associated epigenetic aberrations.
Candidate background
We are particularly interested in receiving applications from candidates with a background in stem cell biology or immunology and molecular biology. Experience with flow cytometry analysis and/or CRISPR/Cas9 technology is desirable. Basic bioinformatics and statistics knowledge and experience is also desirable (e.g. R, Python programming).
Potential Research Placements
- Alex de Mendoza-Soler, School of Biological and Behavioural Sciences, Queen Mary University of London
- TBD, Aim: Experience in clinical aspects of clonal haematopoiesis
- TBD, Aim: Using epigenetic clock and machine learning
References
- Kar, S. P. et al. Genome-wide analyses of 200,453 individuals yield new insights into the causes and consequences of clonal hematopoiesis. Nat Genet, doi:10.1038/s41588-022-01121-z (2022).
- Nachun, D. et al. Clonal hematopoiesis associated with epigenetic aging and clinical outcomes. Aging Cell 20, e13366, doi:10.1111/acel.13366 (2021).
- McKenna, A. et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907, doi:10.1126/science.aaf7907 (2016).
- Ketkar, S. et al. Remethylation of Dnmt3a (-/-) hematopoietic cells is associated with partial correction of gene dysregulation and reduced myeloid skewing. Proc Natl Acad Sci U S A 117, 3123-3134, doi:10.1073/pnas.1918611117 (2020).
- Wu, X. & Zhang, Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet 18, 517- 534, doi:10.1038/nrg.2017.33 (2017).
