Investigating the cancer evolutionary dynamics of complex structural variants and epigenetic alterations
Primary supervisor: Simone Zaccaria, UCL
Secondary supervisor: Özgen Deniz, Queen Mary University of London
Project
Different genetic and epigenetic alterations accumulate in the genome of cancer cells during tumour evolution. To understand this process, several cancer sequencing studies have attempted to reconstruct tumour phylogenesis using somatic mutations or copy-number alterations (CNAs) [1]. While these evolutionary analyses have been useful to reveal some genetic events that drive cancer evolution, mutations and CNAs only represent a fraction of the alterations accumulated in tumours. In particular, epigenetic alterations have been mostly ignored in evolutionary analyses despite their potential key role in cancer progression [2, 3]. A more comprehensive analysis of both genetic and epigenetic alterations is thus important to uncover novel mechanisms of cancer progression and related phenotypes (e.g., treatment resistance).
Different sequencing technologies can be used to identify epigenetic alterations: for example, bisulfite sequencing to identify methylation alterations, and ATAC sequencing to identify alterations in chromosome accessibility. Moreover, the analysis of complex structural variants (SVs) also enables the identification of consequences of epigenetic alterations from DNA sequencing. For example, SVs analysis is used to reveal the insertion of transposable elements (TEs) activated by epigenetic alterations [4], and to enhance the identification of replication-timing alterations (RTAs), which are often associated to alterations in chromosome accessibility [2]. Despite these opportunities, reconstructing tumour phylogenies of epigenetic alterations remains a challenging task because simple evolutionary models for SVs and epigenetic changes cannot be defined.
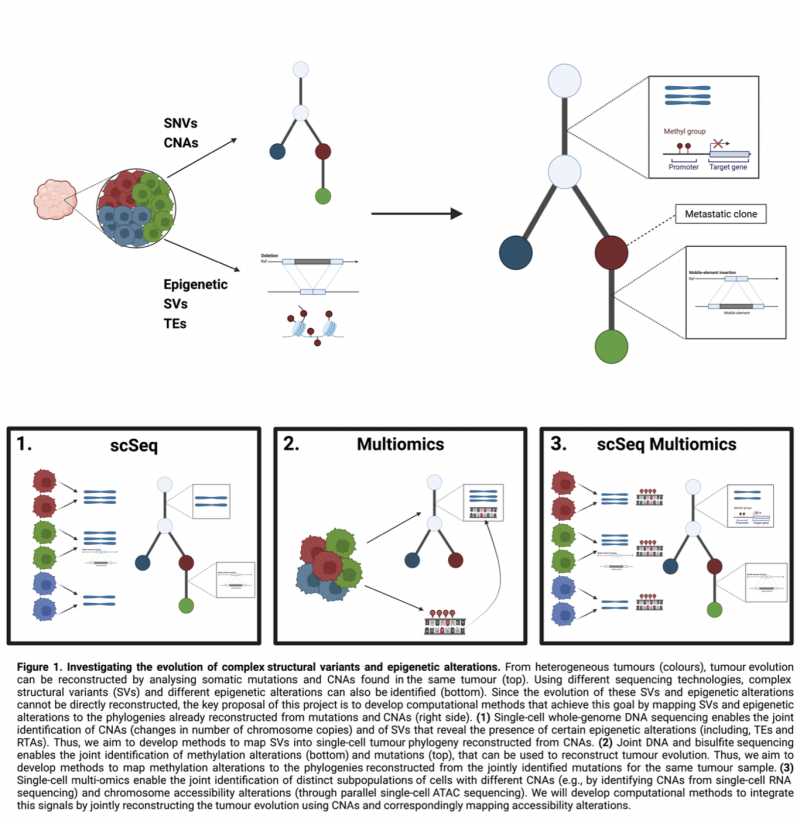
We hypothesise that the reconstruction of their evolution can be enabled by mapping these events into tumour phylogenetic trees that can be inferred in parallel using mutations and CNAs found in the same tumour. Therefore, the goal of this project is to develop and apply computational methods that enable this mapping for SVs and different epigenetic changes (Figure 1). To guarantee the feasibility of the project, we will leverage already existing methods to identify SVs [2,4] and epigenetic alterations [3], and already existing datasets and related phylogenies [1, 3]. The project thus has three key aims.
Aim 1: Reconstructing SV evolution using single-cell sequencing. As part of the PEACE autopsy programme, single-cell whole-genome sequencing data are generated for >100,000 metastatic cancer. Since SVs, TEs, and Investigating the cancer evolutionary dynamics of complex structural variants and epigenetic alterations RTAs can be identified from the same data [2, 4], we will develop methods to map these alterations into the phylogenies already reconstructed using mutations to investigate their role during metastatic progression.
Aim 2: Reconstructing methylation alterations evolution from multi-omics datasets. We will leverage the simultaneous availability of bulk DNA and bisulfite sequencing within the TRACERx non-small cell lung cancer study [3] to reconstruct the evolution of methylation alterations by leveraging already-reconstructed tumour phylogenies.
Aim 3: Reconstructing epigenetic evolution from single-cell multi-omics datasets. The lab of Dr Deniz will be generating single-cell multi-omics datasets. By combining both epigenomics assays and single-cell resolution, we will leverage this setting to develop methods for investigating the role of epigenetic alterations in the evolution of acute myeloid leukaemia, with a special focus on TEs.
Candidate background
This project would suit candidates with a bioinformatics or computer-science background, with expertise in algorithm development and interest in studying cancer evolution.
Potential Research Placements
- Özgen Deniz, Barts Cancer Institute, Queen Mary University of London
- Nnenna Kanu, UCL Cancer Institute
- Benjamin Werner, Barts Cancer Institute, Queen Mary University of London
References
- Frankell, A. M., Dietzen, M., Al Bakir, M., Lim, E. L., Karasaki, T., Ward, S., […] Swanton. The evolution of lung cancer and impact of subclonal selection in TRACERx. Nature, 616, 525-533 (2023).
- Li, Y., Roberts, N. D., Wala, J. A., Shapira, O., Schumacher, S. E., Kumar, K., […] Campbell, P. J. Patterns of somatic structural variation in human cancer genomes. Nature, 578, 7793 (2020).
- Cadieux, E. L., Mensah, N. E., Castignani, C., Tanić, M., Wilson, G. A., Dietzen, M., […] Van Loo, P. Copy number-aware deconvolution of tumor-normal DNA methylation profiles. bioRxiv (2020).
- Rodriguez-Martin, B., Alvarez, E.G., Baez-Ortega, A. et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nature Genetics, 52, 306-319 (2020).
- Zaccaria, S. & Raphael, B.J. Characterizing allele- and haplotype-specific copy numbers in single cells with CHISEL. Nature Biotechnology, 39, 207-214 (2021).
