Unraveling the epigenetic vulnerabilities of recurrent group3 medulloblastoma
Primary supervisor: Laura Donovan, UCL
Secondary supervisor: Roberto Bellelli, Queen Mary University of London
Project
One of the most significant unmet clinical challenges in paediatric oncology is the development of novel therapeutic strategies for recurrent medulloblastoma (R-MB). Following upfront conventional treatments, R-MB occurs in 30% of cases and is near universally fatal, accounting for 10% of all childhood cancer deaths. Standard clinical care at relapse typically focuses on palliation rather than a cure. Routine biopsies of R-MBs have only begun in the last 5-years, as surgery at time of recurrence is associated with significant morbidity; consequently, biological material for interrogation has historically been very rare. Paediatric oncology therapies have commonly been designed using treatment-naïve biopsies but tested in phase I/II trials on children with recurrent disease [1]; unsurprisingly, no successful therapies exist for the treatment of R-MB.
Until recently, assumptions were made that R-MB is identical to the primary tumour and response to therapy will be the same; however, recent evidence suggests that >40% added genetic defects emerge at relapse, with mutations in epigenetic regulators accounting for the majority of genetic pertubations [2,3]. Opportunities for therapeutic exploitation remain poorly understood as the genomics do not yet point to targetable biology. Epigenetic modifications alter DNA accessibility and chromatin structure overtime, regulating patterns of gene expression, without altering the genome sequence itself. This interchangeable nature represents a therapeutically relevant target for primary and R-MB.
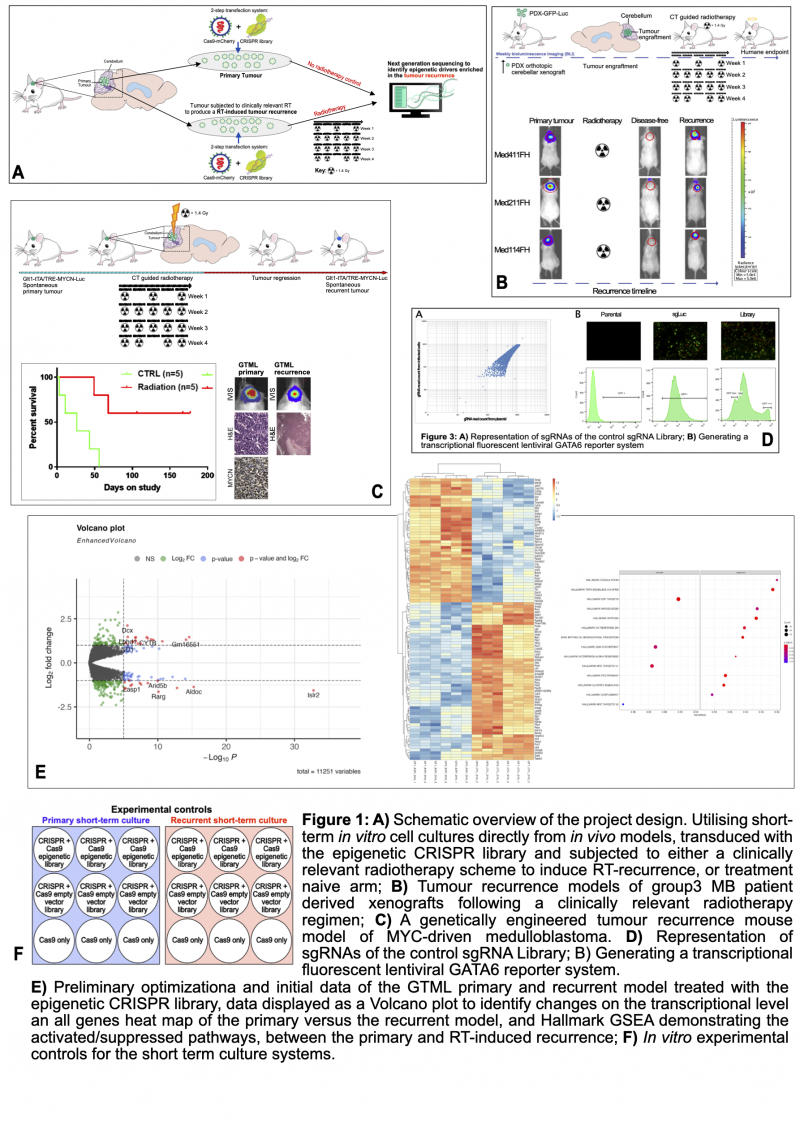
The lead supervisor demonstrated immune priming with epigenetic agents plus immunotherapy as powerful cerebellar tumour therapeutics [4]. We propose to build upon this using epigenetic CRISPR libraries to identify therapeutic vulnerabilities in primary and R-MB models, for validation in immune-competent mouse models for rapid clinical translation (Figure 1A)
We will employ 3-independent patient-derived xenograft MB models (Figure 1B), and a MYC-driven immune-competent MB model (Figure 1C), each proven to faithfully replicate human primary, metastatic, and radiotherapy-induced recurrent disease in vivo, establishing their clinical relevance. Utilizing two epigenetic CRISPR libraries, a mouse library (Figure 1D) (948 genes, 4-gRNAs/gene, totalling 3794-gRNAs) and a human library (Figure 1E) (2508 genes, 5/6-gRNAs/gene, totalling 20,051-gRNAs). These libraries specifically target essential epigenetic driver genes that hold therapeutic potential and have been fully optimized for in vitro applications.
We hypothesise that the dynamics of recurrent medulloblastoma can be mapped using functional genomic CRISPR-Cas9 screens during the exposure to radiotherapy-inducing recurrence in clinically relevant models of MB, to identify genes regulated by epigenetic driver mutations, leading to the identification of targetable therapeutic vulnerabilities.
Aim 1: To conduct loss-of-function epigenetic CRISPR-Cas9 screens in primary versus radiotherapy-induced R-MB (Figure 1A; Figure 1F).
Aim 2: To Identify of candidate epigenetic driver mutations depleted in R-MB for development of a refined epigenetic loss-of-function CRISPR screen.
Aim 3: Validation of candidate driver mutations depleted in the tumour recurrence.
We expect to develop a target list of filtered gene targets depleted in the tumour recurrence, for development into a refined a loss-of-function CRISPR library with single-cell transcriptome resolution [5], for downstream in vivo analysis in clinically relevant immune-competent mouse models of R-MB.
Candidate background
Ideal candidates would have a background in oncology and genetic manipulations (cloning/CRISPR technology/gene knockouts), and an interest in CRISPR-Cas9 technologies, mouse modelling, and bioinformatics.
Potential Research Placements
- Roberto Bellelli, Barts Cancer Institute, Queen Mary University of London
- UCL Genomics, UCL Institute of Child Health
- Louis Chesler, Institute of Cancer Research
References
- Morrissy, A. S. et al. Nature 529, 351?357 (2016).
- Jones, D. T. W. et al. Nature 488, 100-5 (2012).
- Pugh, T. J. et al. Nature 488, 106-110 (2012).
- Donovan, L. K. et al. Nature Medicine 26, 720-731 (2020).
- Donovan, L. K. et al. Nature 572, 67-73 (2019).
