Understanding and exploiting the impact of NPM1 mutations in reshaping the nucleolus of Acute Myeloid Leukaemia
Primary supervisor: Faraz K. Mardakheh, Queen Mary University of London
Secondary supervisor: Paolo Gallipoli, Queen Mary University of London, Elspeth Payne, UCL
Project
Aims:
The aim of this PhD project is to (1) reveal the impact of mutant NPM1 on the composition and interactome of the nucleolus in AML cells, (2) assess the functional consequences of the identified changes on ribosome biogenesis, and (3) evaluate whether any of the identified changes can be therapeutically exploited for treatment of NPM1-mutant AML.
Background:
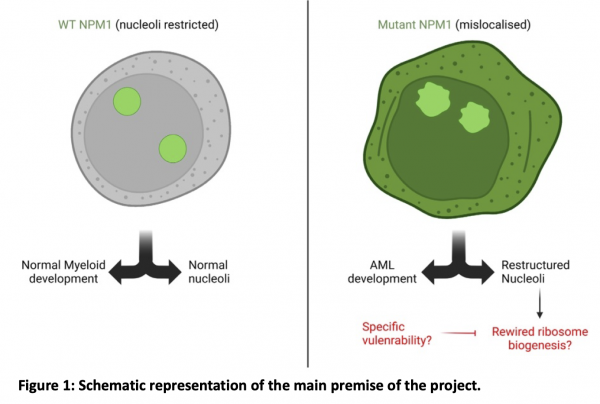
With a five-year survival rate less than 25%, Acute Myeloid Leukaemia (AML) is amongst the most deadly types of blood cancers in adults. The most commonly mutated gene in AML is Nucleophosmin-1 (NPM1), with mutations present in more than 30% of all adult AML cases, and more than half of AML cases with normal karyotype1. NPM1 is an abundant RNA-binding protein (RBP) that is primarily localised to the nucleolus, where it regulates ribosome biogenesis, although it can also shuttle out of the nucleolus to perform additional functions such as chromatin remodelling, DNA damage response, and centrosome duplication. NPM1 mutations in AML are always heterozygous and result in mislocalisation of the mutant proteins to the cytoplasm. It is believed that the mislocalised NPM1 gains novel functions outside of the nucleoli, which are crucial for driving leukaemogenesis. However, the exact molecular mechanisms by which this is achieved is still unclear [1].
Recent studies have highlighted a key structural role for NPM1 in organising the RNA and protein components of the nucleolus [2]. We hypothesize that mislocalisation of NPM1 in AML triggers nucleolar reorganisation, thus rewiring key components of the ribosome biogenesis cascade. As ribosome biogenesis is an essential cellular process that is needed for cell proliferation and viability, a detailed understanding of nucleolar reorganisation in NPM1-mutant AML could reveal novel therapeutic opportunities (Figure 1).
Aim 1 – revealing the impact of mutant NPM1 on the nucleolar composition and interactions in AML cells. Using a previously described isogenic model of NPM1 mutation in AML [3], the student will systematically profile the mutant NPM1-induced changes in nucleolar composition and interactions by quantitative proteomics. For this purpose, they will utilise spatial proteomics as well as RNA-protein interactome capture approaches that are well-established in Mardakheh?s lab [4]. Confocal microscopy and biochemical approaches such as immunoprecipitation will be then employed to verify any detected changes to the composition and interactions, respectively.
Aim 2 – assess the functional consequences of the identified changes on ribosome biogenesis. After revealing the changes in the nucleolar composition and interactions, the student will assess how they can affect the dynamics of ribosome biogenesis. For this purpose, genetic loss/gain of the identified changing factors (e.g. through CRISPR or siRNA depletions vs. overexpression) in the isogenic AML cells with or without NPM1 mutations will be carried out, followed by various cellular and biochemical assays for monitoring various steps of ribosome biogenesis that are established in Mardakheh’s lab (e.g. rRNA synthesis, processing, and ribosome assembly) [4]. The results will be then validated in a panel of primary AML cells, with or without NPM1 mutations. In parallel, the effect of loss/gain of the identified factors on cell proliferation and viability will be assessed, in vitro.
Aim 3 – evaluate whether the identified changes can be therapeutically exploited for treatment of NPM1-mutant AML. The results of Aim 2 may reveal novel dependencies of the NPM1 mutant AML cells to specific ribosome biogenesis factors, revealing opportunities for therapeutic exploitation through synthetic lethality. To assess such possibilities, the student will first use a zebra fish model of normal haematopoiesis [5] (Payne lab) to assess whether targeting any of the identified factors in Aim 2 will significantly compromise normal haematopoiesis. Next, they then use immunocompromised mouse xenografts (Gallipoli lab), to assess the impact of targeting those factors which do not hamper normal haematopoiesis, on AML development, in vivo.
Together, these results will systematically reveal how NPM1 mutations restructure and reorganise the nucleolus, how this reorganisation affects the dynamics of ribosome biogenesis, and whether any of the identified changes have the potential to be therapeutically exploited for AML treatment.
References
- Falini, B., Brunetti, L., Sportoletti, P. & Martelli, M.P. NPM1-mutated acute myeloid leukemia: from bench to bedside. Blood 136, 1707-1721 (2020).
- Mitrea, D.M. et al. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife 5 (2016).
- Brunetti, L. et al. Mutant NPM1 Maintains the Leukemic State through HOX Expression. Cancer Cell 34, 499- 512 e499 (2018).
- Azman, M.S. et al. An RNA-binding switch drives ribosome biogenesis and tumorigenesis downstream of RAS oncogene. bioRxiv, 2021.2012.2016.472890 (2022).
- Otterstrom, J.J., Lubin, A., Payne, E.M. & Paran, Y. Technologies bringing young Zebrafish from a niche field to the limelight. SLAS Technol 27, 109-120 (2022).
