Understanding B Cell Dynamics and ECM Topography in Melanoma Through Multiomic Analysis and Predictive Modelling of Risk Signatures
Primary supervisor: Oscar Maiques, Queen Mary University of London
Secondary supervisor: Sophia Karagiannis, King’s College London
Project
B cells play crucial roles in melanoma immune surveillance and may significantly influence cancer progression and treatment outcomes [1]. Despite their importance, the roles of B cells in melanoma are still not well understood, necessitating further research to elucidate their functions and contributions. Amoeboid cancer cells at the invasive front of melanoma exhibit high-actomyosin contractility and hijack immune cells like macrophages to create a tumour-promoting environment [2,3]. This interaction enhances drug resistance, stemness [4], and tumour progression, highlighting the need for targeted therapeutic strategies. Unpublished work by our group shows that matrix alignment significantly influences melanoma dissemination, with increased alignment and radial orientation at the distal invasive fronts (DIF) compared to proximal fronts or the tumour body [5]. These findings highlight the ECM architecture’s potential as a biomarker for invasion and poor prognosis in advanced melanoma. Furthermore, research in the Karagiannis laboratory has revealed that B cells differentiate, evolve and clonally expand across different compartments of the tumour microenvironment, including tumour islands, stroma and tertiary lymphoid structures (TLS) [1]. Additionally, studying the interactions between B cells and ECM structures may reveal novel biomarkers for predicting risk progression and therapeutic outcomes in melanoma patients.
Description and Aims
We aim to understand the role of B cell dynamics in melanoma progression through a multidisciplinary approach. Initially, by employing Multiplex Cell Dive technology, we will, 1) investigate the relationship between extracellular matrix (ECM) architecture and B cell behaviour during melanoma progression. This will allow us to visualize ECM components and B cell distribution in melanoma tissues, analysing ECM remodelling and correlation with B cell infiltration and identification of TLS (Figure 1A).
Next, 2) utilise spatial transcriptomics data from primary and regional melanoma metastases samples to create detailed signatures of B cell states at the RNA level. This involves identifying and validating B cell-specific gene expression signatures and mapping these to spatially resolved data to understand cell localization and function (Figure 1B), 3) develop deep learning models to predict B cell presence, subtype, and function in histology images using TCGA Skin Cutaneous Melanoma (SKCM) samples. This will involve preprocessing and annotating histology images, training models with bulk RNA-seq data, and validating predictions with independent datasets (Figure 1C). Finally, 4) we will assess the prognostic value of predicted B cell activity in stratifying for overall survival (Figure 1D). A critical aspect of this project is to determine whether ECM architecture or composition contributes to B cell dynamics, particularly TLS formation. This connection could provide significant insights into how ECM remodelling influences B cell behaviour and melanoma progression.
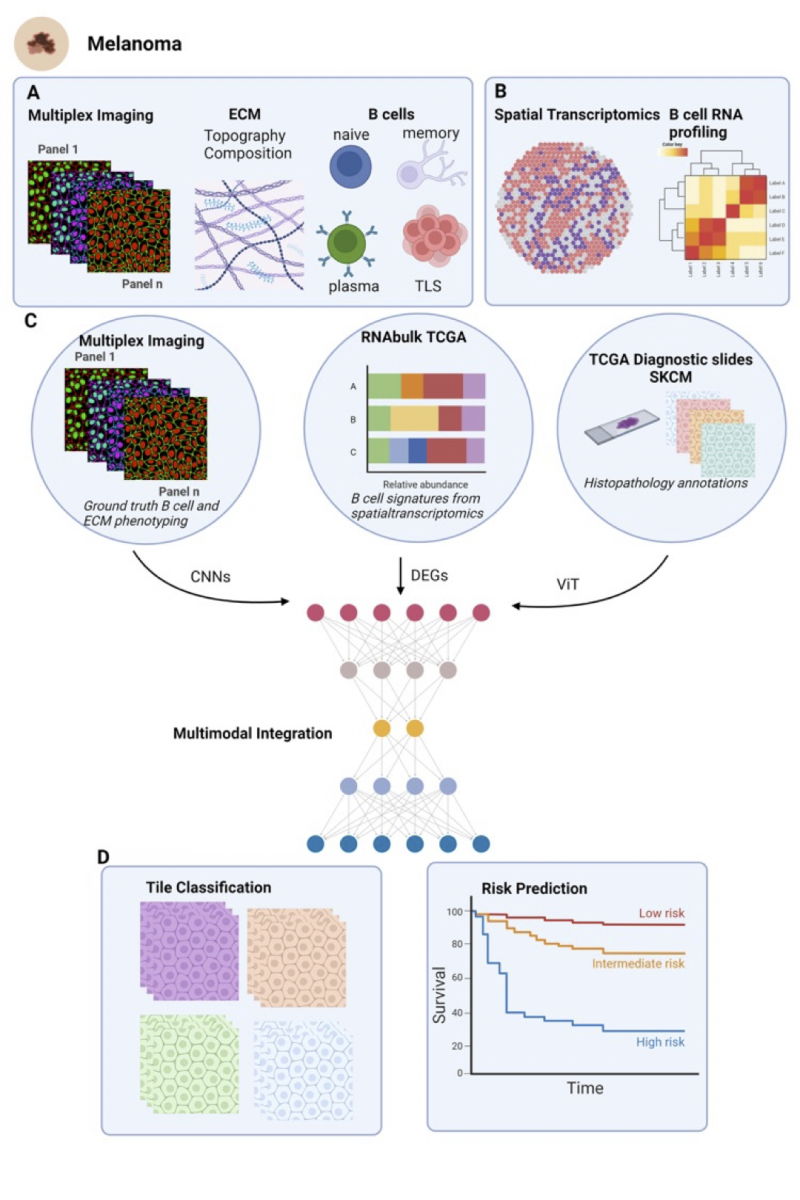
Figure 1: This figure illustrates the workflow of integrating multiplex imaging, spatial transcriptomics, and TCGA data for predicting patient prognosis in melanoma. A) The multiplex imaging panels provide ground truth phenotyping of B cells and ECM composition, capturing various B cell types and their topographical arrangement. B) Spatial transcriptomics identifies B cell markers, which are then used to C) analyze gene expression levels in TCGA RNA-seq bulk data. Histopathology slides from TCGA, annotated for skin cutaneous melanoma (SKCM), are also utilized. Convolutional Neural Networks (CNNs) process the multiplex imaging data, while Vision Transformers (ViTs) analyze the histopathology slides. Differentially expressed genes (DEGs) from the spatial transcriptomics data are integrated into the multimodal model. D) This integrated approach classifies histology tiles based on B cell and ECM activities and predicts patient risk levels, visualized through survival curves indicating low, intermediate, and high-risk prognoses. Created with biorender.
Candidate background
A potential candidate should have a master’s degree in bioinformatics or machine learning. Knowledge in pathology, cancer biology and immunology is recommended. They should possess knowledge of advanced imaging techniques, such as multiplex imaging and segmentation tools, and experience in analysing high-throughput sequencing data, including RNA-seq and spatial transcriptomics (desired). Familiarity with machine learning (ML) and computer vision (CV) domains, particularly deep learning applied to biological datasets, is highly desirable. The candidate should have strong analytical skills, proficiency in programming languages like Python or R.
Potential Research Placements
- Sophia Karagiannis, St John’s Institute of Dermatology, Guy’s Hospital, King’s College London
- Claude Chelala, Barts Cancer Institute, Queen Mary University of London
- Greg Slabaugh, Digital Environment Research Institute (DERI), Queen Mary University of London
References
- Crescioli, S., Correa, I., Ng, J., Willsmore, Z.N., Laddach, R., Chenoweth, A., Chauhan, J., Di Meo, A., Stewart, A., Kalliolia, E., … Karagiannis SN. (2023). B cell profiles, antibody repertoire and reactivity reveal dysregulated responses with autoimmune features in melanoma. Nature Communications 2023 14:1 14, 1–21. https://doi.org/10.1038/s41467-023-39042-y.
- Georgouli, M., Herraiz, C., Crosas-Molist, E., Fanshawe, B., Maiques, O., Perdrix, A., Pandya, P., Rodriguez-Hernandez, I., Ilieva, K.M., Cantelli, G., … Karagiannis SN, Sanz-Moreno V. (2019). Regional Activation of Myosin II in Cancer Cells Drives Tumor Progression via a Secretory Cross-Talk with the Immune Microenvironment. Cell 176. https://doi.org/10.1016/j.cell.2018.12.038.
- Samain, R., Maiques, O., Monger, J., Lam, H., Candido, J., George, S., Ferrari, N., Kohihammer, L., Lunetto, S., Varela, A., et al. (2023). CD73 controls Myosin II–driven invasion, metastasis, and immunosuppression in amoeboid pancreatic cancer cells. Sci Adv 9. https://doi.org/10.1126/SCIADV.ADI0244.
- Rodriguez-Hernandez, I., Maiques, O., Kohlhammer, L., Cantelli, G., Perdrix-Rosell, A., Monger, J., Fanshawe, B., Bridgeman, V.L., Karagiannis, S.N., Penin, R.M., et al. (2020). WNT11-FZD7-DAAM1 signalling supports tumour initiating abilities and melanoma amoeboid invasion. Nat Commun 11. https://doi.org/10.1038/s41467-020-18951-2.
- Maiques O, Sallan MC, Laddach R, Pandya P, Varela A, Crosas-Molist E, Barcelo J, Courbot O, Liu Y, Graziani V, Arafat Y, Sewell J, Rodriguez-Hernandez I, Fanshawe B, Jung-Garcia Y, …, Karagiannis SN, Elosegui-Artola A, Sanz-Moreno V. Matrix mechano-sensing at the invasive front induces a cytoskeletal and transcriptional memory supporting metastasis. Nat Commun. 2025;16(1):1394. doi: 10.1038/s41467-025-56106-w.
