Unravelling molecular determinants of progression and treatment response in NASH-supported hepatocellular carcinoma
Primary supervisor: Gilbert Fruhwirt, King’s College London
Secondary supervisor: Oscar Maiques, Queen Mary University of London
Project
Liver cancers remain a major global health challenge with an estimated incidence >1million/year in 2025 and ~20k male/10k female annual deaths (USA;[Siegel_CACancerJClin_2025]) and ~6k overall in the UK. Hepatocellular carcinoma (HCC) accounts for ~90% of liver cancer cases. Liver damage caused by alcohol misuse and hepatitis B and C virus infections have been the main HCC risk factors, but non-alcoholic steatohepatitis (NASH) associated with metabolic syndrome or diabetes mellitus are now major risk factors in the West and drive incidence increases there (projected ~33% increase in the UK over next 15 years).
Various new treatment options for advanced/unresectable HCC became available over recent decades (immune checkpoint inhibitors (ICI), combination targeted therapy, histotripsy, stereotactic external beam radiotherapy (EBRT)) but 10‑year survival remains only 8% and overcoming therapy resistance is the main challenge. For example, only 15-20% of patients benefit from ICI and 3-year survival after EBRT is only 23%. Hypoxia is a negative predictive factor for response to radiotherapy due to lowering the efficacy of ionizing radiation by a factor of 2-3 [Thoday_Nature_1947;doi:10.1038/160608a0]. Numerous clinical trials investigating combination therapies aim to overcome resistance. However, many mechanistic aspects of the interplay between immune evasion, radiation resistance, and hypoxia remain elusive, particularly in the context of HCC and therein NASH/cirrhosis-induced HCC.
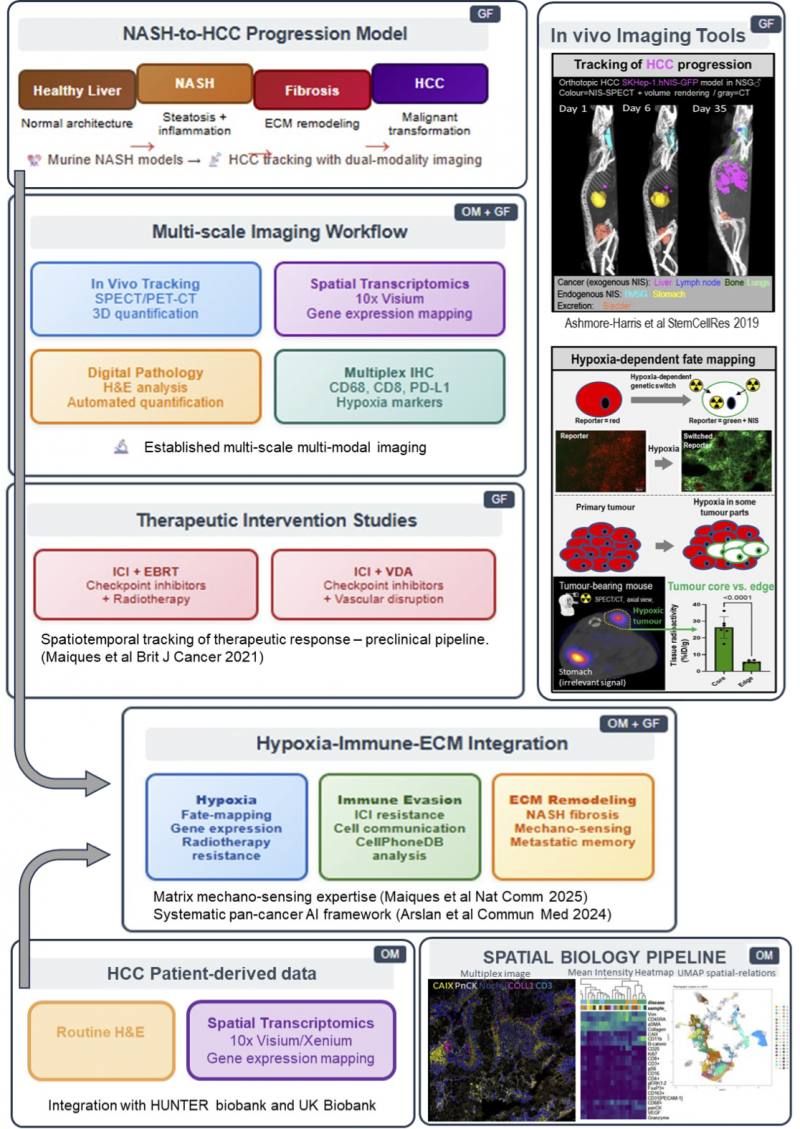
In this project, we aim to define how macroscopic phenomena such as NASH and hypoxia impact on HCC progression and dissemination as well as on HCC immuno- and radiotherapy treatment response on the molecular level. We will harness the combined powers of multi-scale imaging, multi-parametric tissue analyses, and computational data integration (see Figure below) to identify on the molecular level how HCC progresses within liver environments with different underlying NASH statuses.
We build on existing murine NASH models and our established in vivo cancer tracking technology [Fruhwirth_JNucMed_2014, Maiques_BrJCancer_2021] for 3D quantification of cancer growth and metastasis over time and employ high-dimensional spatial tissue analysis methods to investigate HCC progression and treatment response. We will also employ a new in‑house developed tumour hypoxia fate-mapping technology, to investigate how hypoxia alters HCC gene expression profiles in vitro in cells and how it impacts on HCC tumour establishment and progression in vivo. Ex vivo tissue analyses of corresponding animal tumours will reveal how NASH-associated liver fibrosis and hypoxia-driven desmoplasia alter gene expression and extracellular matrix (ECM) architecture, building on our understanding of how ECM mechano-sensing drives cytoskeletal and transcriptional memory supporting tumour aggressiveness [Maiques_NatComm_2025]. We will also challenge HCC tumours with combination therapy (i.e. ICI±vascular disrupting agent or ICI±EBRT) and integrate the resultant spatiotemporal data with observations from human patient-derived data. The latter constitute tissue microarray and clinical outcome datasets from collaborating hepatology centres (e.g. Blizard@QMUL, William-Harvey@King’sCollegeHospital will provide validation cohorts). Integration will employ AI-driven digital pathology pipelines established by Maiques [Arslan_CommMed_2024], using deep learning models to predict multi-omic biomarkers from routine H&E sections, combined with spatial transcriptomics correlation analysis and virtual multiplexing techniques to map progression and treatment response patterns.
Candidate background
Suitable candidates will have a minimum upper 2nd-class degree in cell biology/immunology/biochemistry or a closely related field with a strong background in cancer biology, an interest in advanced multi-scale imaging and multi-parametric analysis, and a passion for hypothesis-driven research. This is a highly multi-disciplinary project, and the student will employ a range of techniques including cell culture, mouse tumour models, diverse imaging technologies (i.e. SPECT/PET-CT, fluorescence microscopy, immunohistochemistry, spatial biology), fluorescence-activated cell sorting, transcriptomics, and computational methodologies for integrated data analysis.
Potential Research Placements
- Anita Grigoriadis, Comprehensive Cancer Centre, King’s College London
- Mirjana Efremova, Barts Cancer Institute, Queen Mary University of London
- Claude Chelala, Barts Cancer Institute, Queen Mary University of London
References
- Siegel RL, Kratzer TB, Giaquinto AN, Sung H, Jemal A. Cancer statistics, 2025. CA Cancer J Clin. 2025;75(1):10-45. doi: 10.3322/caac.21871.
- Fruhwirth GO, Diocou S, Blower PJ, Ng T, Mullen GED. A whole-body dual-modality radionuclide optical strategy for preclinical imaging of metastasis and heterogeneous treatment response in different microenvironments. J Nucl Med. 2014;55(4):686-94. doi: 10.2967/jnumed.113.127480.
- Maiques O, Fanshawe B, Crosas-Molist E, Rodriguez-Hernandez I, Volpe A, Cantelli G, Boehme L, Orgaz JL, Mardakheh FK, Sanz-Moreno V, Fruhwirth GO. A preclinical pipeline to evaluate migrastatics as therapeutic agents in metastatic melanoma. Br J Cancer. 2021;125(5):699-713. doi: 10.1038/s41416-021-01057-y.
- Arslan S, Schmidt J, Bass C, Mehrotra D, Geraldes A, Singhal S, Hense J, Li X, Raharja-Liu P, Maiques O, Kather JN, Pandya P. A systematic pan-cancer study on deep learning-based prediction of multi-omic biomarkers from routine pathology images. Commun Med. 2024;4(1):48. doi: 10.1038/s43856-024-00474-1.
- Maiques O, Sallan MC, Laddach R, Pandya P, Varela A, Crosas-Molist E, Barcelo J, Courbot O, Liu Y, Graziani V, Arafat Y, Sewell J, Rodriguez-Hernandez I, Fanshawe B, Jung-Garcia Y, Imbert PRC, Grasset EM, Albrengues J, Santacana M, Macià A, Tarragona J, Matias-Guiu X, Marti RM, Tsoka S, Gaggioli C, Orgaz JL, Fruhwirth GO, Wallberg F, Betteridge K, Reyes-Aldasoro CC, Haider S, Braun A, Karagiannis SN, Elosegui-Artola A, Sanz-Moreno V. Matrix mechano-sensing at the invasive front induces a cytoskeletal and transcriptional memory supporting metastasis. Nat Commun. 2025;16(1):1394. doi: 10.1038/s41467-025-56106-w.
