Targeting a cell cycle vulnerability in TP53 mutant cancers
Primary supervisor: Tanya Soliman, Queen Mary University of London
Secondary supervisor: Miraz Rahman, King’s College London
Project
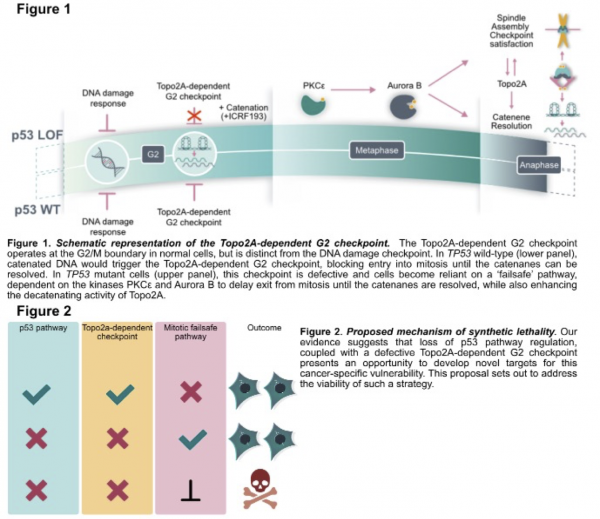
To successfully complete cell division, cells must remove DNA tangles (catenanes) that arise during replication. In normal cells, these tangles would be sensed by a G2-phase cell cycle checkpoint which prevents entry into mitosis until the enzyme, Topoisomerase2A, completes decatenation. We discovered that cancer cells harbouring a TP53 mutation have a defective G2 decatenation checkpoint resulting in impaired cell division and genome instability. To compensate, these p53-mutant cells become reliant on an alternative ‘failsafe’ pathway which delays mitosis and protects from aneuploidy and cell death. We have mapped the molecular details underlying this failsafe pathway: Aurora B, an essential mitotic kinase, changes conformation to access an alternative set of substrates resulting in delayed mitosis and enhanced Topo2A-mediated DNA detangling (Figure 1).
Previous attempts to exploit Aurora B therapeutically in cancer failed to improve patient outcomes in solid tumours and caused severe toxicity, reflecting it essential role in cell division in both normal and malignant tissues. However, our new insight into this specific conformation of Aurora B provide us with a unique angle to revisit the potential to target Aurora B as a cancer therapy. Importantly, our findings identify TP53-mutant cancers (over 50%) as a specific and actionable target for this approach.
We hypothesise that combining a Topo2 catalytic inhibitor (e.g., ICRF193) to induce DNA catenation and activate the mitotic failsafe pathway, with selective targeting of Aurora B, is a potentially viable therapeutic strategy in TP53-mutant cancers (Figure2).
We have developed CL-213-22, a first-in-class allosteric inhibitor that selectively targets a specific conformation of Aurora B kinase. This conformation is induced by PKCε-dependent phosphorylation of Aurora B. Our findings reveal that this phosphorylation event acts as a molecular switch, redirecting Aurora B to an alternative substrate repertoire associated with the mitotic failsafe pathway. CL-213-22 preserves the canonical kinase activity of Aurora B while selectively blocking interactions with substrates involved in the failsafe pathway. Unlike existing Aurora B inhibitors, whether ATP-competitive or allosteric, CL-213-22 does not abolish all kinase activity, instead offers a distinct mechanism-of-action by modulating substrate specificity. This selective inhibition enhances cancer cell targeting while sparing normal cells.
In this project, we will optimise CL-213-22 analogues using high-resolution structural data of the Aurora B–CL-213-22 complex to define the allosteric binding site. Computational methods, including molecular dynamics, docking, and free energy perturbation, will guide rational design. Syntheses will be performed in the Rahman lab, and analogues assessed through biochemical and cellular assays to refine potency, selectivity, and cancer cell-specific activity.
Aims and purpose of proposed investigation: We aim to understand the biological response to p53 pathway dysregulation leading to Topo2A-dependent G2 checkpoint deficiency and determine the molecular requirements for the mitotic failsafe pathway in TP53 mutant cancers. We will explore both the underlying biology and the clinical utility of the novel, allosteric Aurora B inhibitor, CL-213-22.
Aim 1: Determine the precise molecular requirements of the mitotic failsafe pathway
This will involve characterising the biological response to excess metaphase catenation using global and phospho-proteomics to identify signalling networks involved in the mitotic failsafe pathway.
Aim 2: Define the structural basis of selective Aurora B inhibition by CL-213-22
High-resolution structural studies will define the allosteric binding site of CL-213-22 and the conformation of Aurora B associated with Ser227 phosphorylation. These insights will guide the design of analogues with improved binding and selectivity.
Aim 3: Elucidate mitotic failsafe pathway vulnerabilities in TP53 mutant cancer.
Here the student will be seconded to the Rahman lab to develop analogues of CL-213-22 with improved pharmacological profiles. These analogues will be characterised in vitro and in cell lines using established protocols.
Aim 4: Assessment of mitotic failsafe pathway inhibition in cancer models.
Lead candidates of the CL-213-22 analogues will be taken forward for testing in pre-clinical models of TP53 mutant cancer (spheroid, xenograft models)
Candidate background
The project would be suitable for a candidate with a background in biomedical sciences and a keen interest in drug discovery and cancer cell biology. A knowledge of biochemistry or medicinal chemistry would be an advantage but not essential
Potential Research Placements
- Miraz Rahman, Institute of Pharmaceutical Science, King’s College London
- James MacDonnell, Randall Centre for Cell & Molecular Biophysics, King’s College London
- TBC: testing in xenograft/ PDX models
References
- Soliman TN, Keifenheim D, Clarke DJ, Parker PJ. Cell Cycle responses to Topoisomerase II inhibition: Molecular Mechanisms and Clinical Implications. Journal of Cell Biology 222 (12): e202209125. (2023) 10.1083/jcb.202209125
- Lockwood N, Martini S, López-Pardo A, Deiss K, Segeren HA, Semple RK, Collins I, Repana D, Cobbaut M, Soliman TN, Ciccarelli FD, Parker PJ. A distinctive requirement for p53 in the genome protective Topoisomerase 2a-dependent G2 arrest in hTERT positive cancer cells. Cancer Research 82(9): 1762-1773 (2022) 10.1158/0008-5472.CAN-21-1785
- Kelly JR, Martini S, Brownlow N, Jamshidi S, Lockwood N, Joshi D, Federico S, Kjaer S, Rahman KM, Fraternali F, Parker PJ*, Soliman TN*. The Aurora B specificity switch is required to protect from non-disjunction at the metaphase/anaphase transition. Nature Communications 11:1396. (2020) 10.1038/s41467-020-15163-6
- Lewis, T., Corcoran, D. B., Thurston, D. E., Giles, P. J., Ashelford, K., Walsby, E. J., Fegan, C. D., Pepper, A. G., Rahman, K. M., and Pepper, C. (2021) Novel pyrrolobenzodiazepine benzofused hybrid molecules inhibit nuclear factor-κB activity and synergize with bortezomib and ibrutinib in hematologic cancers, haematologica 106, 958.
- Corcoran, D. B., Lewis, T., Nahar, K. S., Jamshidi, S., Fegan, C., Pepper, C., Thurston, D. E., and Rahman, K. M. (2019) Effects of Systematic Shortening of Noncovalent C8 Side Chain on the Cytotoxicity and NF-kappa B Inhibitory Capacity of Pyrrolobenzodiazepines (PBDs), Journal of Medicinal Chemistry 62, 2127-2139.
