Heritability of non-genetic variation in tumour evolution
Primary supervisor: Nicholas McGranahan, UCL
Secondary supervisor: Erik Sahai, Francis Crick Institute
Project
The advent of affordable sequencing has shed light on key driver alterations and mutational processes that sculpt the cancer genome during its evolution (McGranahan et al., 2015). While most work on cancer evolution has focused on genomic events, emerging multi-omic research has highlighted the importance of non-genetic events in influencing response to treatment and likelihood of metastasis (Marjanovic et al., 2020).
However, while non-genetic events may influence the cancer cell phenotype, these can only be subject to selection, including from anti-cancer treatment, if they are heritable (Black & McGranahan, 2021). As such, there is a need to develop approaches to distinguish between heritable and non-heritable diversity in cancer evolution and to explore the impact of these different forms of heterogeneity in response to selection pressures, in particular from therapy.
We propose to explore the importance of heritable and non-heritable diversity as cancers develop and respond to anti-tumour immunity and therapy, aiming to uncover new evolutionary vulnerabilities in cancer which may be therapeutically exploited.
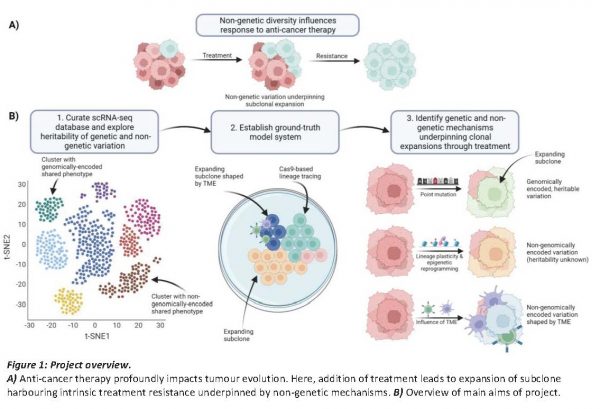
Aim 1. A single-cell database and computational tools for exploration of genetic and non-genetic diversity in cancer
We will collate a compendium of publicly available single-cell RNA sequencing data, downloaded and processed uniformly to study the heritability of tumour-intrinsic non-genetic variation in cancer. Thereafter, computational approaches to assign transcriptional cell states, a proxy for cell phenotype, to cancer cell clones will be developed. The differences between genomically-defined (e.g. by analysis of somatic copy-number alterations or mutations), and transcriptionally-defined cell states (e.g. by expression signatures) will enable an interrogation of heritable and non-heritable diversity.
Aim 2: Establish ground-truth model system to study heritability of cancer cell phenotypes in cancer
In unpublished work, the Sahai group has shown that cancer cells grown on simple 2D substrates represent a system to study clonal expansions and clonal mixing in the context of treatment and microenvironmental pressures in real time. Briefly, this technique will be combined with Cas9-based lineage tracing approaches, and single-cell RNA-sequencing (Quinn et al., 2021) to explore cell states and phenotypic plasticity in the context of clonal dynamics, in the presence or absence of anti-cancer treatment. Further, analysis of cancer cell states in the context of co- cultured microenvironmental cells (Montagner & Sahai, 2020) will allow interrogation of the impact of the tumour microenvironment upon heritable and non-heritable tumour-intrinsic non-genetic diversity.
Aim 3. Identifying genetic and non-genetic mechanisms underpinning subclonal expansions and treatment resistance in cancer
Unpublished analysis from the TRACERx study of lung cancer has revealed that expanded clones show increased signals of positive selection and are more likely to seed metastasis. However, in the majority of such clones, genomically-defined driver events have remained elusive, raising the possibility that many clonal expansions are non-genetically underpinned. In-depth analysis of expanded, and non-expanded clones within the ground-truth model system and TRACERx tumours and their transcriptional phenotypes will enable exploration of non-genetic drivers of cancer evolution, which may be therapeutically exploited. Once again, the link with the Sahai group will enable functional testing of subclonal expansions.
Candidate background
This project would suit candidates with a biological background, strong computational experience, and a desire to pursue a computational project. This project will involve processing multiple publicly available scRNA-seq libraries; experience in designing and running computational pipelines or utilising publicly available datasets would be desirable.
Potential Research Placements
- Eric Sahai, Francis Crick Institute
- James Reading, UCL Cancer Institute
- Simone Zaccaria, UCL Cancer Institute
References
- Black, J.R.M. and McGranahan, N. (2021) ‘Genetic and non-genetic clonal diversity in cancer evolution’, Nature reviews. Cancer, 21(6), pp. 379 – 392.
- Marjanovic, N.D. et al. (2020) ‘Emergence of a High-Plasticity Cell State during Lung Cancer Evolution’, Cancer cell, 38(2), pp. 229 – 246.e13.
- McGranahan, N. et al. (2015) ‘Clonal status of actionable driver events and the timing of mutational processes in cancer evolution’, Science translational medicine, 7(283), p. 283ra54.
- Montagner M, Sahai E. (2020) ‘In vitro Models of Breast Cancer Metastatic Dormancy.’ Front Cell Dev Biol. 3;8:37. doi: 10.3389/fcell.2020.00037.
- Quinn, J.J. et al. (2021) ‘Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts’, Science, 371(6532). doi:10.1126/science.abc1944.
