Cancer recognition code of gamma-delta T-cells
Primary supervisor: Yin Wu, King’s College London
Secondary supervisor: Marco Gerlinger, Queen Mary University of London
Project
Immunotherapy with immune checkpoint inhibitors (ICIs) is highly effective against some cancers but unfortunately many patients do not benefit. ICIs leverage the capacity of “conventional” αβ T-cells to recognise (neo)antigens in the context of self-MHC which leads to specific cancer cell killing. Unfortunately, cancers frequently evade this through defects in antigen presentation. Nonetheless, some of these cancers can still be controlled by the immune system as they demonstrate exquisite sensitivity to ICIs. Several studies have now implicated γδ T-cells as important effectors in these settings [1,2]. These “unconventional” specialist T-cells are enriched within epithelial tissues from which carcinomas arise, and they can detect and kill transformed cells independent of (neo)peptide-MHC restriction [3]. Recent work suggests they are direct effector targets of anti-PD1/PD-L1 ICI therapy [4]. Thus, they represent a potent immunological defence against cancers when conventional T-cells may fall short.
Despite their clear potential in restraining carcinogenesis, little is known about how γδ T-cells function in situ in human tumours. These cells are difficult to isolate from human tissues and most studies to date have either utilised murine models or γδ T-cells expanded from clinical material and manipulated in vitro. Thus, whilst we have some mechanistic insight into how these cells may be regulated, we know little about their true function within clinical tumours. Specifically, how γδ T-cells recognise cancer cells in situ is entirely unknown. Do they employ “adaptive” recognition via their T-cell receptor (TCR)? Do they rely on innate receptors such as NK receptors (NKRs)? Or do they use a mixture of both? An understanding of the in situ modus operandi of γδ T-cells would inform rational strategies to utilise these cells more effectively for therapeutic purposes. For example, TCR-dependent activation of these cells would have implications on responsiveness to anti-PD-(L)1 therapies already in clinical use whilst NKR-dependent activation may require the development of novel therapies targeting this axis.
The advent of robust and high-plex spatial transcriptomics (e.g., NanoString CosMx) now enables the study of these cells in situ within the tumour microenvironment. This project will employ spatial transcriptomics to analyse resection specimens from patients with colorectal cancers, a condition in which γδ T-cells have recently been strongly implicated1, for putative TCR and/or NKR ligands. Additionally, RNA-sequencing, genetic validation screens (e.g., CRISPR) and functional assays using patient-derived tumour models [5] will establish likely molecular mechanisms regulating these cells in clinical disease (Figure 1)
Aims:
- Establish how γδ T-cells recognise cancer cells in resected tumours (via TCR or NKR?).
- Establish the ligands recognised by γδ T-cells in clinical disease and their hierarchical relationship in activating these cells.
- Modulate these molecular switches to improve γδ T-cell cancer immunosurveillance potential.
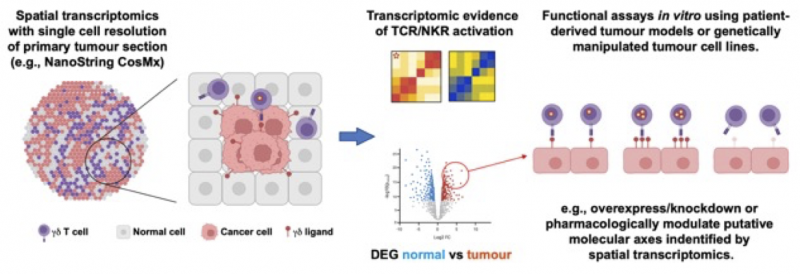
Figure 1. Experimental plan
Candidate background
The candidate must demonstrate enthusiasm for cancer immunology, have a solid grounding in oncology and experience with immunotherapy. Previous wet lab experience, particularly of flow cytometry, molecular biology and primary cell culture are desirable but not essential. Likewise previous experience of coding and biostatistics (e.g., R/Python) is desirable but not essential.
References
- Wu, Y. et al. A local human Vδ1 T cell population is associated with survival in nonsmall-cell lung cancer. Nature Cancer 3, 696-709 (2022).
- Vries, N. L. de et al. γδ T cells are effectors of immunotherapy in cancers with HLA class I defects. Nature 613, 743?750 (2023).
- Mensurado, S. et al. The emerging roles of γδ T cells in cancer immunotherapy. Nature Reviews Clinical Oncology 20, 178-191 (2023).
- Davies, D. et al. PD-1 defines a distinct, functional, tissue-adapted state in Vδ1+ T cells with implications for cancer immunotherapy. Nat. Cancer 1-13 (2024) doi:10.1038/s43018-023-00690-0.
- Gonzalez-Exposito, R. et al. CEA expression heterogeneity and plasticity confer resistance to the CEA-targeting bispecific immunotherapy antibody cibisatamab (CEA-TCB) in patient-derived colorectal cancer organoids. The Journal for ImmunoTherapy of Cancer 7, 101 (2019).
