DDR-Fate: Single-Cell Phenotypic Characterisation of DNADamage Response Dependencies Identified from Whole Genome CRISPR Screens
Primary supervisor: Graeme Hewitt, King’s College London
Secondary supervisor: Chris Tape, UCL
Project
Project background and description:
With the advent of CRISPR-based genetic screens, genome-scale analyses of genegene and gene-treatment interactions have become possible. This has transformed the study of DNA damage repair, allowing researchers to not only identify DNA repair pathways and factors, but also characterise interdependencies and crosstalk between them. This has proved an invaluable approach in identifying novel cancer therapy strategies, as well as understanding and overcoming cancer-treatment resistance (Hewitt et al., Mol Cell, 2021).
Hewitt Lab has recently performed several whole-genome CRISPR screens to map the cellular radiation response. These screens reveal genetic dependencies in relation to various radiation dosing strategies and specific radiation-drug combinations. We have successfully identified most known DNA damage/radiation response pathways but also revealed several novel factors whose role in radiation response is unknown.
While CRISPR screens have greatly accelerated our ability to unravel genetic dependencies, current methods for phenotypic characterisation of the genes identified represents a significant bottleneck. Assays are typically low-throughput and focus on only one or two cellular phenotypes. Consequently, only a few screen hits are followed up with mechanistic analyses, resulting in an underutilization of the information gained from this powerful genetic approach.
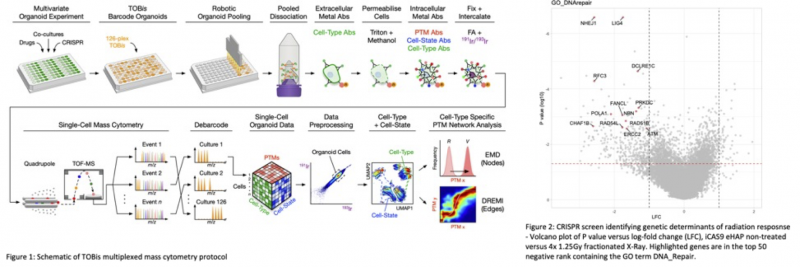
Single-cell technologies such as mass cytometry (MC), allow multiple phenotypes to be measured from the same cell and enable the identification and quantification of complex cell-types and phenotypes. Importantly, because information on >40 proteins and posttranslational modification (PTM) are measured per cell, it is possible to simultaneously map PTM signalling networks, cell-cycle status, apoptosis, and activation of the DNA damage responses. Chris Tape has been at the forefront of developing this technology, building a platform that enables up to 126 single cell comparisons in a single assay (Qin et al., Nature Methods, 2020 and Sufi et al., Nature Protocols, 2021). The existing platform has been recently used to map PTM signalling, cell-state, and cell-fate transitions in >4,000 organoid cultures (Zapatero et al., bioRxiv, 2023, in revision, Cell and Qin et al., bioRxiv, 2023, accepted, Cell) demonstrating its utility for high-throughput phenotypic screening at single-cell resolution (Figure 1).
Aim 1: Develop a custom DDR-Fate MC antibody panel to assess cell cycle, DNA Damage Response (DDR) and cell fate. Tape lab have extensive experience of constructing and validating antibody panels for use with MC. This project will utilise suitable validated markers from existing panels and expand these to include a comprehensive set or markers for: cell cycle, DDR, replication, apoptosis, necrosis, autophagy, senescence and innate immunity.
Aim 2: Characterise response to genotoxic agents using DDR-Fate panel. The DDR-Fate panel will be used to characterise cellular response to specific genotoxins E.g. replication stress generated with hydroxy urea (HU) treatment. Characterisation of our panel in response to defined damage will allow us to benchmark against existing assays as well as construct phenotypic signatures that correspond to specific cellular responses e.g. replication stress, double strand break (DSB) repair etc.. The signatures generated in this aim will inform on the repair processes that might be affected when assessing uncharacterised genes in Aim 3.
Aim 3: Characterise hits from radiation response CRISPR screens using DDR-Fate panel. Hits will be selected from radiation response screens in an unbiased manor, based on defined thresholds for log fold change and p-value from MaGeck comparisons (figure 2). Inducible knockout cell lines will be generated by lentiviral introduction of gRNAs targeting selected hits into eHap and Hela Kyoto cells expressing DOX-inducible Cas9. Cell fate phenotypes will be assessed +/- X-ray irradiation using the DDR-Fate panel following gene knockout. 126-plex single cell comparison will allow the high-throughput characterisation of multiple combinations of genotypes and timepoints at single-cell resolution. This will result in a rich mechanistic dataset which closely describes the interplay between genotype and phenotype in radiation response.
This approach will allow characterisation of hits that would not be accessible using current methodology, expanding the utility of genetic screens, and increasing the chances of identifying disease relevant mechanisms and targets.
Candidate background
This project would suit a candidate with a background in life sciences, an interest in genome stability and the application of emerging technologies. An affinity for computational analysis is also desirable.
Potential Research Placements
- Chris Tape, UCL Cancer Institute
- Anita Grigoriadis, Comprehensive Cancer Centre, King’s College London
- Roberto Bellelli, Barts Cancer Institute, Queen Mary University of London
References
- Hewic, G. et al. Defecdve ALC1 nucleosome remodeling confers PARPi sensidzadon and synthedc lethality with HRD. Mol Cell 81, 767-783 e711, doi:10.1016/j.molcel.2020.12.006 (2021).
- Qin, X. et al. Cell-type-specific signaling networks in heterocellular organoids. Nat Methods 17, 335-342, doi:10.1038/s41592-020-0737-8 (2020).
- Sufi, J. et al. Muldplexed single-cell analysis of organoid signaling networks. Nat Protoc 16, 4897-4918, doi:10.1038/s41596-021-00603-4 (2021).
- Zapatero, M. R. et al. Trellis Single-Cell Screening Reveals Stromal Reguladon of Padent-Derived Organoid Drug Responses. bioRxiv, 2022.2010.2019.512668, doi:10.1101/2022.10.19.512668 (2023).
- Qin, X. et al. A Single-cell Perturbadon Landscape of Colonic Stem Cell Polarisadon. bioRxiv, 2023.2002.2015.528008, doi:10.1101/2023.02.15.528008 (2023).
