How does the PKCα-D463H oncogenic mutation drive epigenetic reprogramming and cancer?
Primary supervisor: Angus Cameron, Queen Mary University of London
Secondary supervisor: Neil McDonald, Francis Crick Institute
Project
A single D463H point mutation to the PRKCA gene (encoding protein kinase C-α; PKCα) is the primary driver of chordoid glioma, a rare brain tumour with few treatment options beyond complex surgery. We have shown that heterozygous knock-in of this protein kinase inactivating D463H mutant to the endogenous PRKCA locus in mice is also oncogenic, driving early onset chondrosarcoma and early mortality [1]. How does this single heterozygous point-mutation drive oncogenic transformation?
PKCa is a Ser/Thr protein kinase known to regulate cellular processes including proliferation and apoptosis. We have shown that the D463H mutation is kinase inactivating but also acts as a dominant gain-of-function oncogenic mutant, operating through an allosteric mechanism analogous to gain-of-function small GTPase mutations which retain unhydrolyzed GTP (Figure 1A). This concurs with our previous work showing glioma dependence on PKCα protein but not kinase activity [2,3]. Evidence suggests that PKCα-D463H promotes epigenetic reprogramming to drive transformation through chromatin regulating components, including BRD4 and HCF-1. This work can define novel mechanisms regulating tumour cell plasticity. More broadly, allosteric kinase regulation, analogous to small GTPase switches, can have paradigm shifting significance for the therapeutic targeting of kinases.
Project Plan:
Aim 1: Molecular basis for PKCα-D463H gain-of-function mechanism. Expression of the D463H mutants drives remodelling of the glioma transcriptome and PKCα interactome (Figure 1B). Enhanced interaction with BRD4 and MLL1 complex components, coupled with Myc pathway transcriptional upregulation (Figure 1C) suggest epigenetic oncogenic reprogramming. Intriguingly, alterations to the interactome are ATP dependent because a related D463N mutant, which shows poor ATP retention behaves differently, with a distinct interactome and no oncogenic properties in vivo [1]. Preferential interactors with D463H will be explored using: (i) Cell biology: co-localisation, pull-downs, functional assays; (ii) structural work elucidating binary and higher-order complexes between D463H mutant and identified partners using single particle analysis of cryo-electron microscopy data; (iii) In silico modelling of PKC allostery and putative target interactions (Fraternali collaboration).
Aim 2: Exploring how PKCa-D463H drives epigenetic reprogramming. We have shown that the D463H decreases glioma cell sensitivity to two distinct BET domain targeting inhibitors, indicative of epigenetic reprogramming (Figure 1D-G). ATAC-seq and CUT&RUN analyses will explore changes in chromatin binding and PKCa, BDR4, HCF-1 promoter occupancy (Ficz collaboration). They will also explore changes to specific chromatin marks associated with epigenetic targets identified, including H3K4 and H3K27. This work will explain how PKCa-D463H can promote cellular re-wiring and transformation.
Aim 3: Beyond PKC: atypical behaviour and allosteric regulation of protein kinases. The student will take a bioinformatic approach to identify kinases, which may show differential functional outputs depending on GTPase-like allosteric switching. Somatic mutations to kinase domain functional residues will be explored in pan-cancer datasets for potential gain-of-function mutants. In silico modelling, nucleotide occupancy predictions and biochemical validation will explore kinases of interest.
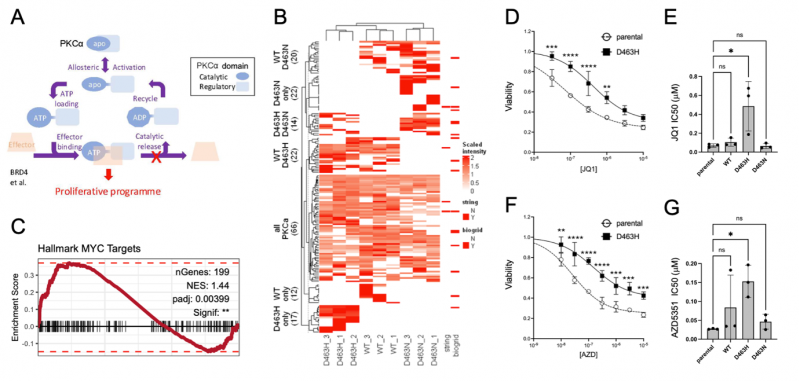
Figure 1: Model of PKCα-D463H gain-of-function mechanism and preliminary data. A. D463H mutation locks PKCα in an ATP bound state to drive allosteric effector activation. B. The D463H mutation has a distinct protein interactome to both WT-PKCα and the non-ATP binding D463N mutant. C. D463H drives activation of Myc target genes and (D-E) decreases glioma cell sensitivity to BET inhibitors.
Candidate background
The supervisory team have a strong collaborative track record examining protein kinase function and allostery for PKC and EGFR family members [1-5]. This project would suit candidates with interests in cell and structural biology, epigenetics, in silico modelling and cancer.
Potential Research Placements
- Neil McDonald, Francis Crick Institute
- Franca Fraternali, Institute for Structural & Molecular Biology, UCL/ Birkbeck
- Gabriella Ficz, Barts Cancer Institute, Queen Mary University of London
References
- Calleja, V., J. C. Henry, M. Cobbaut, J. Sewell, K. Rizzoti, F. Houghton, S. Boeing, N. Anyanwu, S. Varsani-Brown, T. Snoeks, A. Suárez-Bonnet, S. L. Priestnall, N. Q. McDonald*, Cameron AJM* and P. J. Parker (2025). The penetrant chordoid glioma PRKCA mutation is an oncogenic gain-of-function kinase inactivation eliciting early onset chondrosarcoma in mice. bioRxiv: 2025.2005.2028.656646. Science Signaling. IN PRESS. *Co-senior and corresponding authors
- Cameron AJM, Escribano C, Saurin AT, Kostelecky B, Parker PJ. PKC maturation is promoted by nucleotide pocket occupation independently of intrinsic kinase activity. Nat Struct Mol Biol. 2009 May; 16(6): 624-30. (Commentary: Frye SV, Johnson GL. Inhibitors paradoxically prime kinases. Nat Chem Biol. 2009 Jul;5(7):448-9.
- Cameron AJM, Procyk KJ, Leitges M, Parker PJ. PKC alpha protein but not kinase activity is critical for glioma cell proliferation and survival. Int J Cancer. 2008 Aug 15;123(4):769-79.
- Earl CP, Cobbaut M, Barros-Carvalho A, Ivanova ME, Briggs DC, Morais-de-Sá E, Parker PJ, McDonald, N.Q. (2025) Capture, mutual inhibition and release mechanism for aPKC-Par6 and its multi-site polarity substrate Lgl. Nat Struc Mol Biol. doi: 10.1038/s41594-024-01425-0. PMID: 39762628. (Commentary: see News and Views. A PAR6-aPKC-LGL structure reveals how LGL antagonizes aPKC. St Johnston D. Nat Struc Mol Biol. PMID: 40016343)
- ClausJ, Patel G, Autore F, Colomba A, Weitsman G, Soliman TN, Roberts S, Zanetti-Domingues LC, Hirsch M, Collu F, George R, Ortiz-Zapater E, Barber PR, Vojnovic B, Yarden Y, Martin-Fernandez ML, *CameronAJM, Fraternali F, Ng T, Parker PJ. Inhibitor-induced HER2-HER3 heterodimerisation promotes proliferation through a novel dimer interface. Elife. 2018 May 1;7. pii: e32271. *Co-senior and corresponding author.
