Characterisation of metabolic heterogeneity and evolution in IDH-mutant AML in response to targeted therapies
Primary supervisor: Lynn Quek, King’s College London
Secondary supervisor: Paolo Gallipoli, Queen Mary University of London
Project
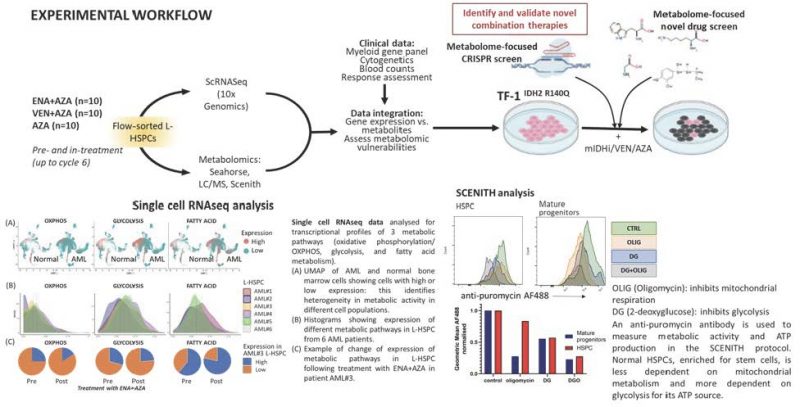
Acute myeloid leukaemia (AML) has a 5-year survival rates of only ~30%. Heterogeneity in mutations, gene expression and metabolic profiles are shaped by therapeutic pressures and drive resistance (1, 2). Leukaemic haematopoietic stem-progenitor cells (L-HSPC) have distinct metabolic features from normal HSPC, that confer competitive advantage and sustain their increased bioenergetic needs. However, we need to better understand how metabolism differs between L-HSPC in patients undergoing treatment. AML mutated in isocitrate dehydrogenase (mIDH) 1 and 2 (~20% of AMLs) are an ideal model to study rewired metabolism. MIDH1/2 inhibitors including Enasidenib (ENA, a mIDH2 inhibitor) are approved to treat IDH-mutant AML, alone or in combination with hypomethylating agents (e.g. azacytidine, AZA) (3). AZA is also combined with Venetoclax (VEN, a BCL2 inhibitor) to treat AML4. With both regimens, although most patients respond, the majority then relapse. Furthermore, ~20-30% of patients are refractory to these treatments.
We already have scRNAseq of L-HSPC from mIDH2 AML patients before and in-treatment with ENA+AZA (n=10) and AZA alone (n=5), from which we identified multiple distinct L-HSPC populations in individual patients. These L-HSPCs express specific metabolic pathways at different stages of treatment and indicate inter- and intra-patient heterogeneity in L-HSPC phenotypes pre-treatment which evolve during therapy. We hypothesise that such metabolic states are likely drivers of leukaemogenesis which can be actioned therapeutically and yield biomarkers of response to current or future therapies.
Aims
- Compare transcriptomic profiles of mIDH patients treated with ENA+AZA, VEN+AZA and AZA alone: obtain scRNAseq data on L-HSPC from mIDH patients, pre- and in-treatment with VEN+AZA (n=10), and a further 5 patients treated with AZA alone. Analyse these data with existing RNAseq data on patients treated with ENA+AZA.
Platforms: Illumina NGS, 10x Genomics Chromium
Expertise: Quek Group (KCL) - Metabolomic profiling of L-HSPCs on longitudinal samples from mIDH2 patients treated with ENA+AZA (n=10) and VEN+AZA (n=10) compared with AZA alone (n=10). We will choose responders and non-responders patients to correlate findings with outcome.
Platforms: Seahorse metabolic flux analysis, Liquid Chromatography/ Mass Spectrometry, SCENITH single cell flow cytometric metabolic profiling (5).
Expertise: Gallipoli group (Barts), Maria Yuneva and Richard Burt (Crick). - Integrate metabolomic data with RNAseq data to understand how changes to gene expression of metabolic genes relate to changes in relevant metabolites.
Expertise: Quek and Gallipoli groups - Perform a metabolic-focused CRISPR/ drug screen in a mIDH2 AML cell line (TF-1IDH2R140Q) to assess synthetic lethality with ENA, AZA and VEN to provide orthogonal validation of pathways identified in Aims 1-3 and identify therapeutically actionable metabolic targets.
Expertise: Quek and Gallipoli groups, and collaboration with Prof JJ Schuringa in Groeningen, Netherlands.
Facilities and resources:
Equipment for flow cytometry cell sorting and metabolomic profiling, and expertise in analysis of RNAseq and metabolome data are established in the Quek and Gallipoli groups. Patient samples have been sourced from the AML-005 study (NCT02677922), and the Haematology Biobanks (KCL:REC#18/NE/0141, Barts:REC#21/EE/0123). We will use CoLCC core facilities for Single Cell Genomics and the HPC Centre.
Candidate background
The innovative aspect of this project is the integration of longitudinal genomic and metabolomic data in patient AML samples to generate new insights in dysmetabolism which has potential for drug target discovery. There is a clear translational line of sight and is ideal for a clinician scientist interested in cancer multiomics.
References
- Quek, L. et al. Clonal heterogeneity of acute myeloid leukemia treated with the IDH2 inhibitor enasidenib. Nat. Med. 24, 1167-1177 (2018).
- Gallipoli, P. et al. Glutaminolysis is a metabolic dependency in FLT3ITD acute myeloid leukemia unmasked by FLT3 tyrosine kinase inhibition. Blood 131, 1639-1653 (2018).
- DiNardo, C. D. et al. Enasidenib plus azacitidine versus azacitidine alone in patients with newly diagnosed, mutant-IDH2 acute myeloid leukaemia (AG221-AML-005): a single-arm, phase 1b and randomised, phase 2 trial. The lancet oncology 22, 1597-1608 (2021).
- Pollyea, D. A. et al. Impact of Venetoclax and Azacitidine in Treatment-Naïve Patients with Acute Myeloid Leukemia and IDH1/2 Mutations. Clinical cancer research 28, 2753-2761 (2022).
- Argüello, R. J. et al. SCENITH: A Flow Cytometry-Based Method to Functionally Profile Energy Metabolism with Single-Cell Resolution. Cell metabolism 32, 1063-1075.e7 (2020).
